Evolutionary origins of honey bees
Who you buzzin′ round now Billy bee, Billy bee
Who you buzzin′ round now?
Why you buzzin′ round her not me?
The Haden Triplets
The honey bee genus Apis includes at least a dozen species. Among these, only the western honey bee (Apis mellifera) occurs in Europe. More than 30 subspecies are found there and in Africa and the Near and Middle East (Table 1 in Carr, 2023). Honey gathering is depicted in petroglyphs from the Iberian Peninsula 8,000 years ago, and apiculture is well documented in Egypt as far back as 2600BCE. From their Old World origins, western honey bees have been transported around the world as a source of honey and beeswax, and in the Americas as pollinators of vegetable and fruit crops that have also been imported from elsewhere. Geographic variation among different local subspecies of A. mellifera is known to contribute to the quality and quantity of honey produced, their use as pollinators and other behaviours.
There is thus a great deal of practical interest in understanding the genetic variation and evolutionary history of honey bees. But first an admission: apart from a single course in entomology as a biology undergraduate, I know almost nothing about bees and beekeeping. My specialisation is genetics and evolution, and I want to tell this story as an example of a typical scientific investigation. This is what I found.
Morphological studies of honey bee subspecies have recognised four continental groups, designated ACMO: African, Continental (European), Mellifera, and Oriental (Near and Middle Eastern). The nucDNA (explained below) genome agreed broadly with this arrangement and suggested an ‘Out of Africa’ hypothesis (Diagram 3) with two invasions of Europe and Asia from Africa (plus ‘Africanised’ bees in the New World). An alternative ‘Out of Asia’ hypothesis looked at the same data, but proposed invasions of Europe and Africa from Asia.
How it’s done
What would I find if I started over from scratch? First, I have to tell you about a particular DNA molecule, mitochondrial DNA (mtDNA), a small, circular molecule found outside the cell nucleus in the power-generating mitochondria, where its dozen or so genes contribute to cellular metabolism. Unlike biparentally inherited nuclear DNA (nucDNA), the mtDNA genome is inherited solely through the maternal line: mothers pass it to all their offspring, but only the daughters pass it to their offspring, and so on. MtDNA was the first DNA molecule to be used in the reconstruction of evolutionary trees and, as it evolves much more rapidly than nucDNA, it is very good at tracing evolutionary lineages over shorter evolutionary times. It’s worked well in my own studies of human populations, other mammals, fish and the occasional sea monster (tinyurl.com/BC2023-12-01).
As a scientific editor, I was asked to review a paper on the use of variation in short mtDNA molecules to identify honey bees. To check the authors’ conclusions, I began to make a more complete meta-analysis, an assembly of data from previous authors into a single, comprehensive analysis. To that end, I compiled the complete mtDNA genome sequences of nearly 100 individual bees from nine species of Apis, including 22 subspecies of A. mellifera. This is just short of one million DNA base pairs (ACGT letters) (Diagram 1).
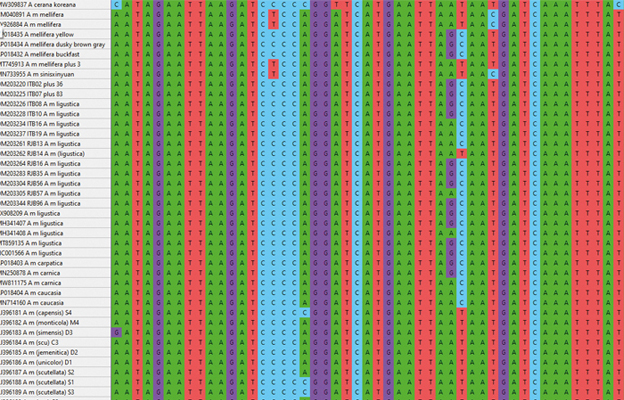
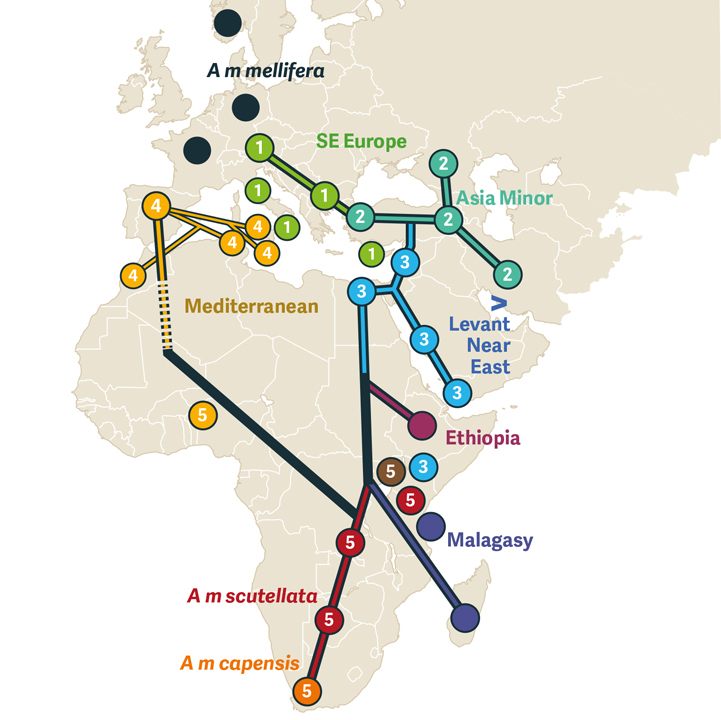
Diagram 2. Phylogeographic evolution of western honey bees as inferred from mtDNA data. Redrawn from Figure 3 of Carr (2023).
Numbered symbols summarise five clades described in the text. Dark and light green circles indicate respectively subspecies in the 1 Southeast European and 2 Asia Minor clades as part of the Eurasian superclade. Blue symbols 3 indicate the Near and Middle Eastern subspecies from the Levantine, Nilotic, and Arabian coast. Light and dark purple circles indicate independent Ethiopian and Malagasy subspecies, respectively. Light orange symbols 4 indicate subspecies in the Mediterranean clade. Circles numbered 5 in various colours indicate the various Sub-Saharan subspecies.
Phylogenetics, the use of genetic data (such as mtDNA genome sequences) to reconstruct the evolutionary history (phylogeny) of species, can be based on different assumptions and models about how DNA evolves. They all start by identifying all nucleotide changes in the ACGT codes among individuals, subspecies and species: these are called single-nucleotide polymorphisms (SNPs) or snips.
The data for this meta-analysis come from an online database of DNA sequences, GenBank, which includes all DNA sequences published in the scientific literature. The genus Apis can be seen in the Taxonomy Database: tinyurl.com/BC2023-12-02. Like museum collections of pinned insects and skinned animals, GenBank is curated. Entries for individual DNA sequences (accessions) include a lot of molecular information. Does it code for a protein? Where do the coding regions start and stop? How are multiple genes in a single mtDNA genome organised? Also included is information about the source of the specimen. For example, curation for GenBank accession KY926884.1, my reference sequence of A. m. mellifera, includes as below.
/organism = “Apis mellifera mellifera”
/sub_species = “mellifera”
/specimen_voucher = “1410”
/country = “Norway”
/collected_by = “Prof. F. Ruttner”
/note = “stored at Ruttner Bee Collection at the Bee Research Institute at Oberursel, Germany”
Much like the record for a specimen in a museum case, this links that specimen with the molecular data.
The analysis proceeded in three stages. First, I used the mtDNA of a bumblebee (Bombus ignites) to sort out the phylogeny of nine species of Apis honey bees. I used several methods of reconstruction (see Panel). This confirmed previous studies that the so-called cavity-nesting species including A. mellifera and A. cerana (the Indian honey bee) are most recently evolved from the dwarf and giant honey bees, and that A. mellifera is the earliest group to diverge from the other cavity nesters.
Next, I made some trial runs with just a few specimens and subspecies of A. mellifera at a time, with A. cerana to sort out relationships.
The third stage was to assemble all the mtDNA genomes from every subspecies available (22) with as many replicate sequences from different individuals in the same subspecies (80) into a single evolutionary tree (see Figure 2 of Carr 2023).
Things Fall Apart with Out of Africa versus a Scramble for Africa
The meta-analysis offers a radically new molecular phylogeography (genetic relationships in their geographic context) for subspecies of A. mellifera (Diagram 2). The ancestral A. m. mellifera originated in northern Europe, diversified in Southeastern Europe, expanded into Asia Minor. European bees then spread southward via the Levant into the Nile Valley and across the Red Sea down the Yemeni coast of the Arabian Peninsula. Southward expansion left distinct Ethiopian and insular Malagasy subspecies. Sub-Saharan African forms are part of a single lineage. Mediterranean honey bees are a secondary return, first to the Iberian Peninsula and thence to the western islands and coastal African oases.
Divergence times among animal taxa can be estimated from a molecular clock, based on the observation that molecular differences and time of divergence are closely correlated. Use of a standard calibration for insect mtDNA suggests that the genus Apis diversified in the late Miocene (c16 million years ago). A. mellifera diverged from other cavity-nesters during the late Pleistocene (c800,000 years ago) at about the same time as Neanderthals diverged from anatomically modern humans. Separation of Euro-African lineages from Sub-Saharan lineages was as recently as 250,000 years ago. Diversity in the Mediterranean group may have been influenced by the most recent Ice Age, c20,000 years ago.
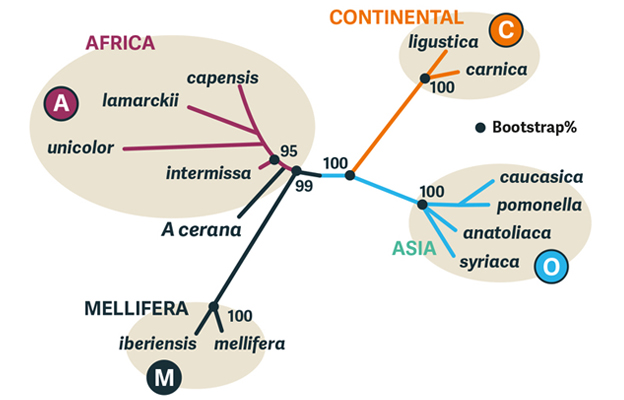
Diagram 3. Out of Africa model of Apis mellifera evolution. Analysis of nucDNA genome data recovers the same four groups (ACMO) as conventional morphology and arranges them as (MA + OC). Use of the Indian honey bee A. cerana as the closest relative places the root of the tree in the African clade (a group with a common ancestor). An alternative Out of Asia model (Han et al 2012) places the root closer to the base of the Asian clade, on the branch between the MA and OC pairs. Compare with Diagram 4 for geographic origins of named subspecies. (Redrawn from Han et al (2012) after Whitfield et al 2006.)
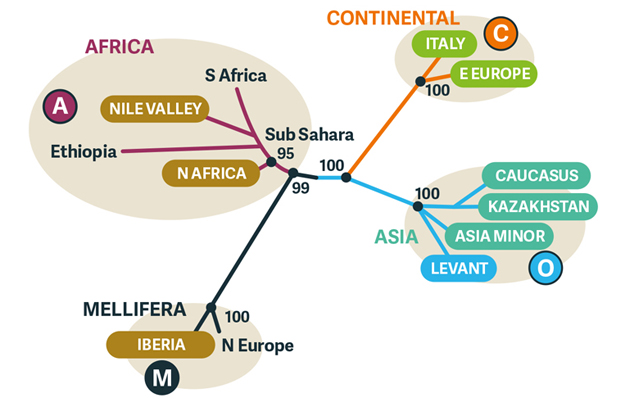
Diagram 4. Comparison of nucDNA Out of Africa/Asia versus mtDNA Scramble for Africa phylogeography. Compare with subspecies names in Diagram 3. Some of the principal differences are separation of the southeast European and Near Eastern clades in the Eurasian superclade (green), separation of the Levantine and Nile Valley clades (blue), and separation of the North African and Iberian clades by joining the latter back to A.m.mellifera.
Some of the main differences between the Out of Africa and the Scramble for Africa hypotheses are summarised in Diagram 4. No MA+OC (Mellifera-Africa + Asia Minor-Continental) model places European bees as basal to African subspecies, whereas the mtDNA data place the latter as the latest products of A. mellifera evolution. The MA+OC model scatters closely related, geographically contiguous subspecies within the Mediterranean mtDNA clade (a group with a common ancestor), including Iberian and North African oasis forms, over the M, A and C clusters.
Similarly, the continuous distribution of Syrian, Yemeni, and Nile Valley subspecies in the mtDNA data are dispersed over the A (Africa) and O (Asia Minor and Caucasian) clusters. On the other hand, groupings of C (Continental European) and O subspecies are similar in both models.
The MA+OC model tends to support the classic morphological picture as a separation among continents. The mtDNA-based model instead describes successive movement across and between continents that are of multiple origin, such that honey bees in Europe, Asia (Near and Middle Eastern) and Africa are of multiple origin.
What to make of these differences? Contrasts between nucDNA, mtDNA and even morphological phylogenies are not unknown. Hybridisation between adjacent subspecies of honey bees may offer an explanation. A queen of one subspecies, arriving in a new area, where she is courted by drones of a different subspecies, will combine her mtDNA with the nucDNA of both species. Recruitment of her hybrid offspring as new queens, with her mtDNA and mixed nucDNA, can allow the mtDNA to jump the subspecies boundary if successive generations continue to backcross to local drones. North African queens newly arrived in Iberia may offer a model. There are some cool experiments here.
“You’ve no idea how confusing it is ….”
”I don’t think they play at all fairly,” Alice began, in rather a complaining tone, “and they all quarrel so dreadfully one can’t hear oneself speak – and they don’t seem to have any rules in particular: at least, if there are, nobody attends to them – and you’ve no idea how confusing it is all the things being alive.”
Lewis Carroll’s Alice’s Adventures in Wonderland
Conclusions:
Working melittologists (beekeepers) may take home some general conclusions about genetics.
1. “I don’t know anything about this, but I’ll tell you what I think.” Meta-analysis is a useful method of rechecking previous conclusions as new data become available. Methods of molecular systematics are the same across taxa. Traditional taxon-specific knowledge remains important and together with increasingly sophisticated molecular approaches may lead to crucial new insights.
2. “Get a second opinion.” Many of the Apis subspecies examined are represented by a single specimen. Duplicates begin to reveal more complex patterns and even errors. DNA sequencing used to be difficult and time-consuming. Now that it’s much easier, identifications should be checked by multiple specimens.
3. “Don’t believe everything you read.” Consultation of the original literature on honey bee systematics as well as the GenBank database should be done with a critical eye. Systematics evolves.
4. “Names Matter.” Even in the 21st century, so-called ‘Alpha Taxonomy’, the basic science of recognising different forms of creatures, remains important, even in well-studied taxa like Apis.
5. “DNA does not lie.” When I began my graduate work in genetics, knowing the complete ACGT code of any complex species was an impossibly far-off dream. A few decades later, this is a routine operation of a few days work for a few thousand dollars. The newly emerging science of bioinformatics requires proper curation, improved methods of information science and novel approaches to analysis in order to understand and make efficient use of the vast amounts of data now available.
How it’s done
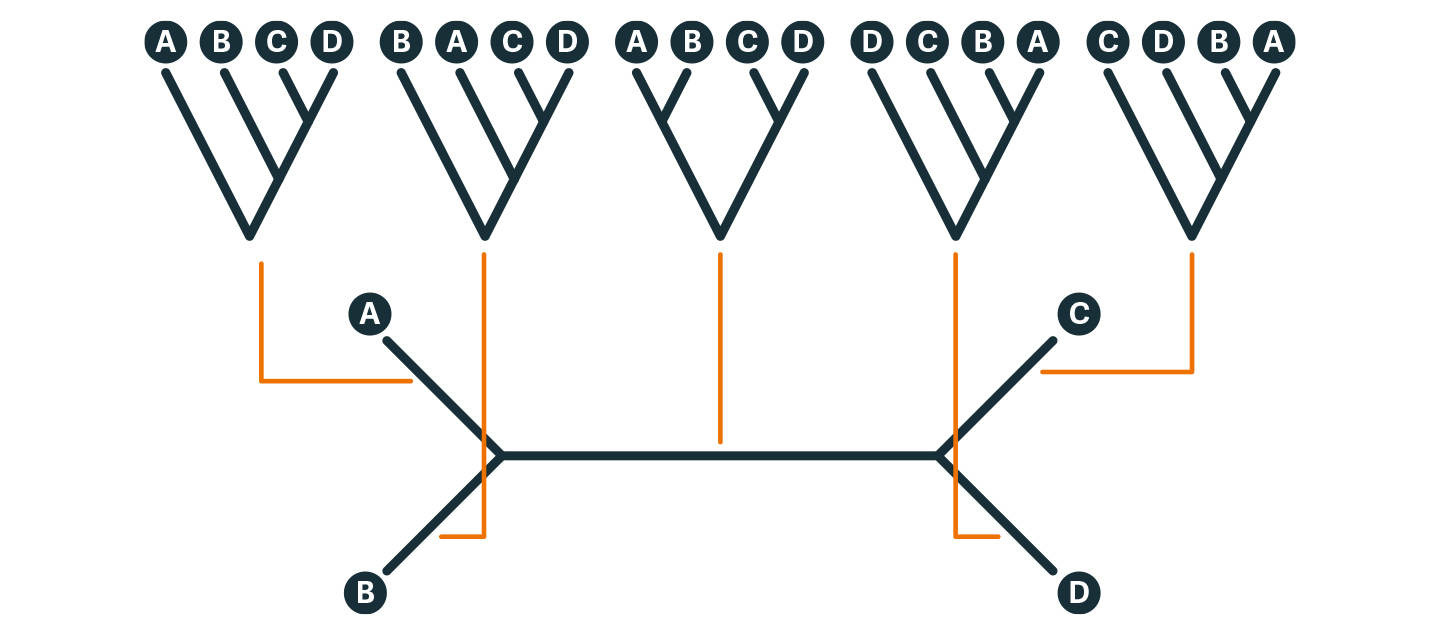
Phylogenetic systematics is a well-established numerical method of approaching taxonomic questions in a rigorous, mathematical manner. The ACMO model of honey bee evolution provides an example of what is called the four taxon problem. Call the taxa A B C D (Diagram 5). We first ask which pairs are most closely related to each other. This gives us a network. To turn the network into an evolutionary tree, we add a root based on an outgroup less closely related to A B C D than they are to each other.
Answering a larger evolutionary question, such as the one discussed here, uses the same logic but requires a lot more decisions. There is a variety of computer algorithms (a set of instructions to solve a problem) that approach the problem in different ways; I used three.
After lining up the DNA sequences, neighbour-joining counts the number of SNP differences between every pair as a genetic distance, and calculates the shortest distance network among all taxa simultaneously.
Maximum parsimony considers the pattern of SNP differences at each site, and calculates the minimum number of changes necessary to get that result. The solution that requires the least number of changes over all sites is the simplest, most parsimonious solution.
Maximum likelihood starts with a precise model of how SNP changes occur: the model allows calculation of the combined probability of all changes necessary to produce any given tree. Although any particular tree is extremely unlikely, one is much less unlikely (maximally likely) than the multitude of all possible trees. It therefore has the maximum likelihood.
Diagram 5 (above). Inferring an evolutionary tree from molecular data: the Four Taxon Problem
With four named taxa A B C D, there are exactly three possible relationships: A is most closely related to B, or to C, or to D, and shown as an H-diagram. Suppose A and B are more closely related, as shown. To infer their evolutionary relationships, we need to add a ‘root’ to the network to make it a ‘tree’. One way to do this is add another taxon, an ‘outgroup’ known to be more distantly related to A B C D than they are to each other. Placement of this outgroup will determine which of the five possible trees (rooted with red lines) is correct, which will in turn clarify the evolutionary relationships of the ‘ingroup’ taxa to each other. For example, the middle tree indicates two pairs of taxa, similar to MA + OC in the Out of Africa model. A + B and C + D, similar to separation of the A+M and C+O honey bee groups in Diagram 2.
References
Carr SM (2023). Multiple mitogenomes indicate Things Fall Apart with Out of Africa or Asia hypotheses for the phylogeographic evolution of Honey Bees (Apis mellifera). Science Reports 13: 9386 – 9398.
Han F, Wallberg A, Webster MT (2012). From where did the Western honeybee (Apis mellifera) originate? Ecology and Evolution 2 (8): 1949 – 1957
Whitfield CW, Susanta K, Behura SK, Berlocher SH, Clark AG, Johnston JS, Sheppard WS, Smith DR, Suarez AV, Weaver D, Tsutsui ND (2006). Thrice out of Africa: ancient and recent expansions of the honey bee, Apis mellifera. Science 314 (5799): 642–645.

Dr Steven Carr
Dr Steve Carr received his PhD in Genetics from UC Berkeley. He is professor of Biology at the Memorial University of Newfoundland in St John’s, Canada, with interests in the molecular phylogeography of fish and mammals in the Nearctic and North Atlantic. Together with the Miawpukek [Mi’kmaq] First Nation, he is investigating genetic continuity between ancient and modern indigenous peoples of Newfoundland.


